
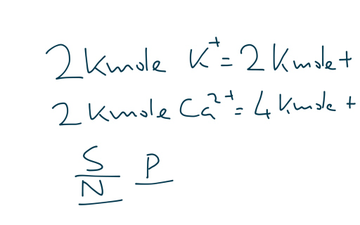
Many selection queries can be strung together using the and or or operators and grouped with brackets. Logical operators can be is, not, >, <, ≥, or ≤. All the IDs and names are as per conventions of the MM engine, i.e., the GROMACS topology. The syntax for the selection query involves the following general structure: where atom selection can include resname for the residue name, resid for residue ID, name for the atom name, type for the atom type, id for the atom ID, and mol for the molecule/chain. (25) It includes selections by atom/residue properties grouped by Boolean operators. This is done by using a custom selection language provided by MiMiCPy, which is designed to be human readable similar to the ones offered by CHARMM, (26) VMD, (24) and PyMOL. Launching the mimicpy prepqm command starts an interactive session, where the atoms to be included in the QM region can be selected. Currently, MiMiCPy supports GROMACS topology (. The minimal set of arguments to be passed to the PrepQM subcommand are the topology and coordinate files. However, the accessible time scales in QM/MM molecular dynamics (MD) simulation are currently limited, especially when applying first-principles methods like density functional theory, (9) which in turn affect the sampling accuracy of QM/MM MD. (8) These methods offer an excellent trade-off between accuracy and computational cost.
(5−7) Here, the system is split into a relatively small QM subsystem and a larger MM subsystem, which are treated at different levels of theory either by different programs (loose-coupling scheme) or within the same program (tight-coupling scheme). (2−4) Currently, the most accurate and computationally expedient way to describe these processes is provided by hybrid quantum mechanics/molecular mechanics (QM/MM) multiscale approaches. (1) These include enzymatic reactions, photobiological processes, and transition-metal ion binding to biomolecules. Biochemical processes span a wide range of time and length scales and often require explicit modeling of electronic degrees of freedom.


 0 kommentar(er)
0 kommentar(er)
